Translational Control of Cell Cycle and Differentiation

ICREA Research Professor
Research information
Background
Recent years have seen a paradigm shift in our understanding of gene activity and regulation. It is now clear that processing of primary transcripts as well as translational control open a myriad of opportunities for gene regulation, which are extensively used in virtually every human gene. However, how these events are regulated and how alterations of these finely tuned processes contribute to physio/pathological processes is not yet well understood.
The primary interest of our group has been to understand the molecular mechanisms that dictate alternative 3’ UTR formation and the temporal and spatial translational control of specific mRNAs during cell cycle progression and chromosome segregation, senescence and related pathologies. Cell cycle progression is programmed, at least in part, by stored silent mRNAs. These mRNAs are not translated en masse at any one time, or even at any one place; rather, their translation is specifically regulated by sequences located at their 3´-untranslated regions (3´-UTRs) and their binding proteins.
Research lines
Our work focuses on four main lines of research:
First, to elucidate the mechanisms underlying the translational control by cytoplasmic polyadenylation cis-acting elements and trans-acting factors:
1) Genome-wide identification of the mRNAs that are regulated by nuclear and cytoplasmic changes in their poly(A) tail length;
2) Determination of the configuration of cis-acting elements that define the temporal and spatial translational regulation;
3) Functional and structural characterization of the ribonucleoprotein (RNPs) complexes that mediate this translational regulation.
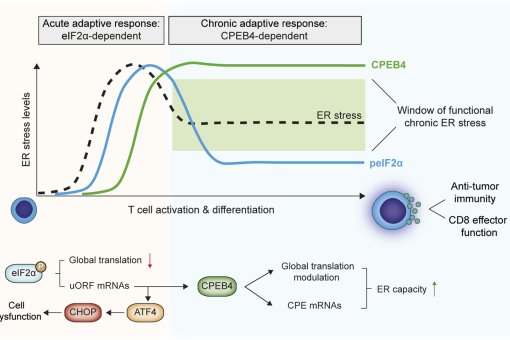
Second, to obtain insights in how this translational control circuit regulates cell cycle progression by establishing a molecular circuit, stabilized by positive and negative feed-back loops to generate an irreversible self sustain hysteric system with molecular memory and switch-like phase transitions.
Third, to explore the role of these mRNA processing and translation mechanisms in the reprogramming of gene expression in tumoral events and angiogenesis and the development of tools with prognostic and therapeutic value.
Forth, to study the symmetric distribution of cellular components (research directed by Oriol Gallego, see www.gallegolab.org)
1)A comparative analysis of Multisubunit Tethering Complexes
2)Characterize the interplay between the transport of vesicles and mRNA localization
Selected publications
Projects
Grup de Recerca consolidat Translational Control of Cell Cycle and Differentiation funded by the Agencia de Gestió d'Ajuts Universitaris i de Recrerca (AGAUR) 2014 SGR 0127

"Estudio combinatorial de los circuitos de expresión génica establecidos por la familia CPEB de proteinas de unión a RNA: mecanismos deacción y nuevas funciones celulares" funded by Ministerio de Economia y Competitividad and Fondo Europeo de Desarrollo Regional (FEDER) de la Unión Europea. BFU2014-54122-P

"Molecular regulation of the progression from hepatic steatosis to cirrhosis and hepatocellular carcinoma: Role and therapeutic potential of CPEB proteins" funded by Fundación Científica Asociación Española contra el Cancer (AECC) GCB15152955MÉND

"Role and therapeutic potential of CPEB proteins in the transition from cirrhosis to hepatocellular carcinoma" funded by Worldwide Cancer Research (WCR) 16-0026

Grup de Recerca consolidat (SGR 2017-2019) Secretaria d'Universitats i Recerca del Departament d'Empresa i Coneixement de la Generalitat de Catalunya. Agencia de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). Reference: 2017 SGR 1451

"Regulación traduccional de la división celular, la diferenciación y el cancer: análisis in vivo de las funciones y mecanismos de acción de las CPEBS" funded by Ministerio de Ciencia e Innovación - Agencia Estatal de Investigación and Fondo Europeo de Desarrollo Regional (FEDER) de la Unión Europea. BFU2017-83561-P

"Metabolic reprogramming in obesity-drven liver cancer, mechanisms, functions and therapeutic potential" AYUDAS FUNDACIÓN BBVA A EQUIPOS DE INVESTIGACIÓN CIENTÍFICA 2018
"Normalization of tumor vasculature through pharmacological inhibition of p38a: Effect on chemotherapeutic drug delivery" funded by Fundació Bancaria Caixa d'Estalvis i Pensions (La Caixa) LCF/PR/HR19/52160020

"Reprogramació metabòlica del càncer de fetge basat en l'obesitat: mecanismes, funcions i oportunitats terapèutiques" funded by Fundació La Marató TV3. 201926-30

"Targeting CPEB-mediated translational balance in Hepatocellular Carcinoma" funded by Worldwide Cancer Research (WCR) 20-0284

"Control traduccional mediado por CPEBs en la interacción tumor-nicho: Una perspectiva mecanística" funded by Ministerio de Ciencia e Innovación - Agencia Estatal de Investigación PID2020-
"Desarrollo de inhibidores de CPEB con propiedades antitumorales" funded by the project PDC2021-121716-I00, funded by MCIN/AEI/10.13039/501100011033 and by the European Union “NextGenerationEU”/PRTR”