The mechanism unveiled triggers a mutation fog, causing hundreds of mutations in each tumor, which spread through the genome of lung, head-and-neck and breast cancers.
Researchers from the Genome Data Science Lab have identified the antiviral APOBEC3A enzyme as the major cause of this new type of hypermutation.
Published in Nature Genetics, the study shows how the mutation fog process generates many oncogenic “cancer driver” mutations, thus accelerating tumour development.
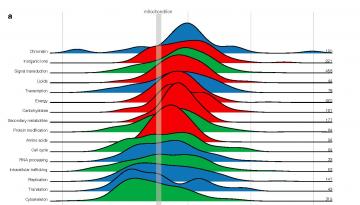
Hypermutation is an unusual occurence that can lead to many nearby mutations at once, severely damaging our genetic material and potentially causing cancer. The best known type of local hypermutation, called a mutation shower or thunderstorm, is quite uncommon and it leads to many mutations accumulated in a small area, e.g. a single gene.
Researchers from IRB Barcelona’s Genome Data Science Lab, led by the ICREA researcher Fran Supek, have discovered a new type of hypermutation called mutation fog, which can generate hundreds of mutations in every cell. Such mutations are widely distributed, but accumulate in the most important regions of the genome, where genes reside (the so-called euchromatin). The fact that these mutations are spread around explains why they have remained undetected until now.
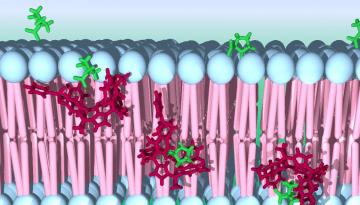
Surprisingly, the scientists also identified that the newly discovered hypermutation type is related to a normal DNA repair process. When cells sense a mismatch in their DNA, they undergo a DNA repair reaction, in order to preserve genetic information. Remarkably, this reaction can become coupled to the APOBEC enzyme–typically used by human cells to defend against viruses and having an important role in fighting hepatitis and HIV. The work by the Genome Data Science Lab indicates that, in some cases, when both the APOBEC enzymes and the DNA repair process are active at the same time, APOBEC hijacks the DNA repair, generating the mutation fog.
“We think that this APOBEC-driven mutation fog has a mutagenic potential that matches or even exceeds that of well-known strong carcinogens, such as tobacco smoke or ultraviolet radiation,” Dr. Fran Supek explained. Recent work by other research groups suggests that the process appears to be more active in late-stage metastatic cancers: it helps the cancer evolve, enabling it to resist drugs and radiation. “This finding makes APOBEC an attractive target for treating cancer, removing its ability to evolve and to become more aggressive,” added Dr. Supek.
The origin of a half of the mutations in some lung and breast cancers
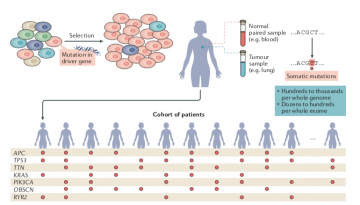
A thorough analysis of more than 6,000 human cancer genomes, including lung tumours, breast tumours and melanomas, among others, led to the finding that the mutation fog is a common phenomenon. “More than half of all APOBEC mutations in some lung or breast cancers are generated by the hypermutation mechanism that we have found,” said David Mas-Ponte, first author of the study and PhD student in the Genome Data Lab.
Some types of cancer, such as cervical or some head-and-neck cancers, are known to be due to viruses. However, this study has found mutations caused by this APOBEC system not only in these tumours but also in cancers that are not currently known to be virus-related. Further work should clarify what triggers the APOBEC system. “Understanding APOBEC better could have broad implications for cancer treatment,” added Mas-Ponte.
The HyperClust statistical method
Mas-Ponte and Supek designed a statistical method, called HyperClust, that can rapidly analyse large amounts of human genomic data to find unusual mutational processes that can lead to simultaneous mutations, such as these cases of mutation fog. This statistical method is described in the article, published in Nature Genetics, and is also available as an open-source software in a Github repository.
This work was funded by the ERC Starting Grant “HYPER-INSIGHT” awarded to Dr. Fran Supek; ICREA reaearcher and EMBO Young Investigator; and the Severo Ochoa grant awarded to IRB Barcelona. David Mas-Ponte was the recipient of an FPI-SO fellowship.
Reference article:
David Mas-Ponte & Fran Supek
DNA mismatch repair promotes APOBEC3-mediated diffuse hypermutation in human cancers
Nature Genetics (2020) DOI: 10.1038/s41588-020-0674-6