
The mechanism unveiled triggers a mutation fog, causing hundreds of mutations in each tumor, which spread through the genome of lung, head-and-neck and breast cancers.
Researchers from the Genome Data Science Lab have identified the antiviral APOBEC3A enzyme as the major cause of this new type of hypermutation.
Published in Nature Genetics, the study shows how the mutation fog process generates many oncogenic “cancer driver” mutations, thus accelerating tumour development.
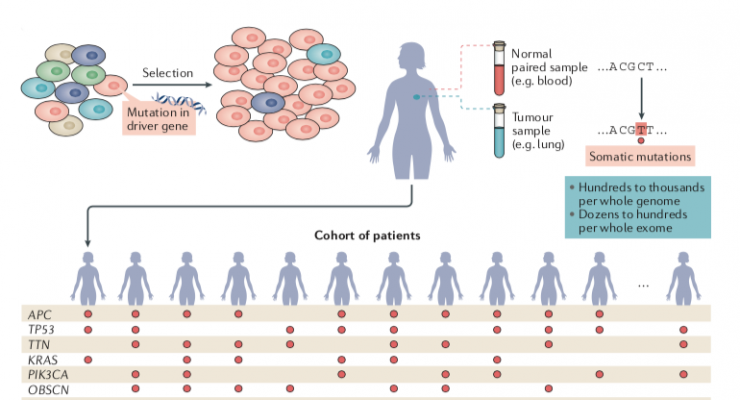
Analysis of the genomes of 28,000 tumours from 66 types of cancer led to the identification of 568 cancer driver genes
Performed by the Biomedical Genomics Lab at IRB Barcelona, the study allowed a major update of the Integrative OncoGenomics (IntOGen) platform, aimed at identifying mutational cancer driver genes.
Published in Nature Reviews Cancer, the results provide the most complete snapshot of the compendium of cancer driver genes to date.