Very close to the Dead Sea there is a valley known as Qumran. In 1947, a set of nine hundred scrolls of written in Hebrew, Aramaic and Greek were found in eleven caves separated by only one kilometer. This discovery stirred a great deal of interest as these manuscripts contained parts of the Old Testament. Today they are known as the Dead Sea Scrolls. Until very recently there were several theories about their origin, the most widespread being that some Jewish sects had kept these scrolls hidden away for a long time.
For many years, the letters used and other physical features of the manuscripts were studied with the aim to unravel their origin. But less than two years ago, they began to be studied in a very different way. In this regard, in June 2020, a very interesting article was published in a scientific journal; researchers had managed to reconstruct the puzzle of these manuscripts by decoding the DNA of the animal skins on which the texts had been written.
The puzzle was very difficult. In total there were about 25,000 fragments of lambskin. Each manuscript could be made from a number of lambskins. A long text may have needed several skins. In addition, the organic treatment to which the skin was subjected in order to make it usable as parchment could affect the genomic analysis performed. Analysis of ancient DNA is not easy and has certain technical complications.
However, a group of researchers specialising in genome analysis took up this challenge and achieved some very interesting results. By studying the genome of the lambskin pieces they were able to determine which ones belonged to the same animal. Thus, these researchers, whom we might refer to as genomic detectives, faced challenges that resemble puzzles and they solved them by analyzing the genome of the parchments.
In the Biomedical Genomics group at IRB Barcelona, we also act like archaeologists or genomic detectives, as there is also a part of our research in which we look for clues that will give us the information we need to study cancer. This part of our work focuses on determining the footprints that are left in tumours by the processes that lead to cancer, and thus discovering the origin and history of each tumour. Using this approach, we define what researchers call "Mutational Footprints" (more technically also called "Mutational Signatures"). These are patterns of mutations that can give us a lot of information about the specific type of tumour we can find.
Each distinct mutagenic process gives a particular characteristic to the pattern of mutations, we call it the "footprint" of mutations or "Mutational Signature".
A well-known footprint is the one left by exposure to the chemical components contained in tobacco. This means that in cancers caused by tobacco smoke, a very specific pattern of mutations is often found that is related to the DNA damage processes produced by the benzopyrenes present in tobacco.
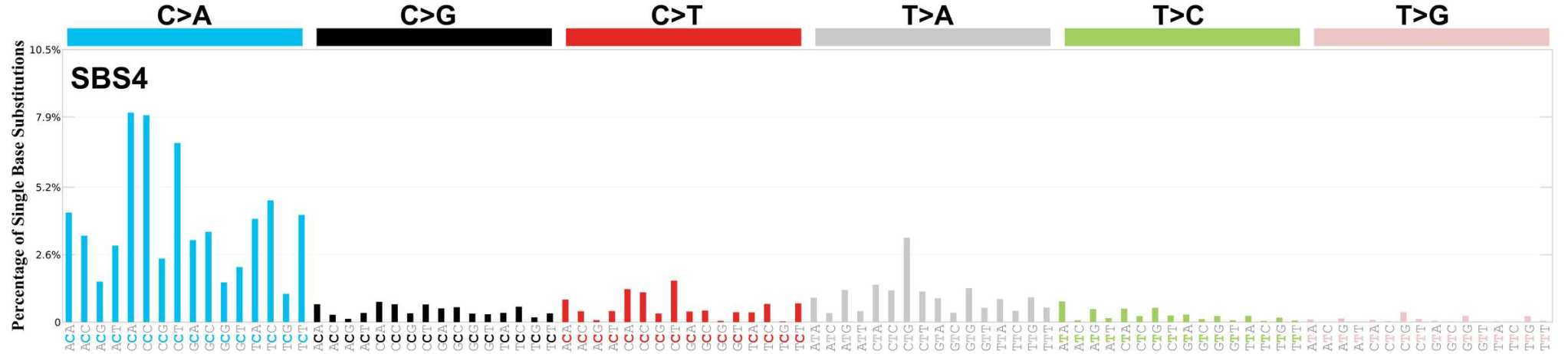
Other "footprints" that we address are those that some cancer treatments themselves can cause. Chemotherapy, in parallel with its antitumour effects, can lead to mutations in originally healthy tissues and this can cause long-term side effects. If we can analyze the patterns of mutations triggered by these chemical compounds and study their effects, then we can better understand which treatments and which doses are best suited to treat cancer and also what their effects may be in the future, thus trying to minimize their side effects.
Our experience as “genomic detectives” has also allowed us to collaborate on a project “tracing” the genome of SARS-COV2 (the virus that everyone knows causes COVID-19). Accustomed to using mathematical models and bioinformatics tools to analyse the mutational processes that occur in cancer, this work was a question of applying all this knowledge to unravel the mutational processes that occur during the spread of the virus.
The study of the genome in different contexts, Biomedical Genomics can be used in studies related to human history or to combat the virus that has recently determined the way we live. But in our case, above all, it helps us to study a group of diseases under the umbrella term of cancer, which at the moment also affect a large part of the population.
On the basis of data from the IARC (International Agency for Research on Cancer), it was estimated that by 2020 there would be around 19.3 million new cases of cancer worldwide*.
Cancer has not suddenly appeared like the SARS-COV2 coronavirus, but by 2020 there were more than 10 million cancer deaths worldwide (according to the same sources). Knowledge of the mechanisms associated with this disease can help us in the treatment and diagnosis of this condition, and most importantly, can help us to improve patients’ quality of life.
Links to the articles mentioned in the post:
Illuminating Genetic Mysteries of the Dead Sea Scrolls
The mutational footprints of cancer therapies
Extracted from the blog "La Ciència a l'ADN" by Martina Gasull. Published with permission of its author.