ICREA Research Professor, UPF Full Professor
Meet Our Scientists Videos
Research information
Background
Our research is focused on the study of cancer from a genomics perspective. We are particularly interested in the identification of cancer driver mutations, genes and pathways across tumour types and in the study of their potential as therapeutic targets.
Research lines
1. Understanding mutational processes
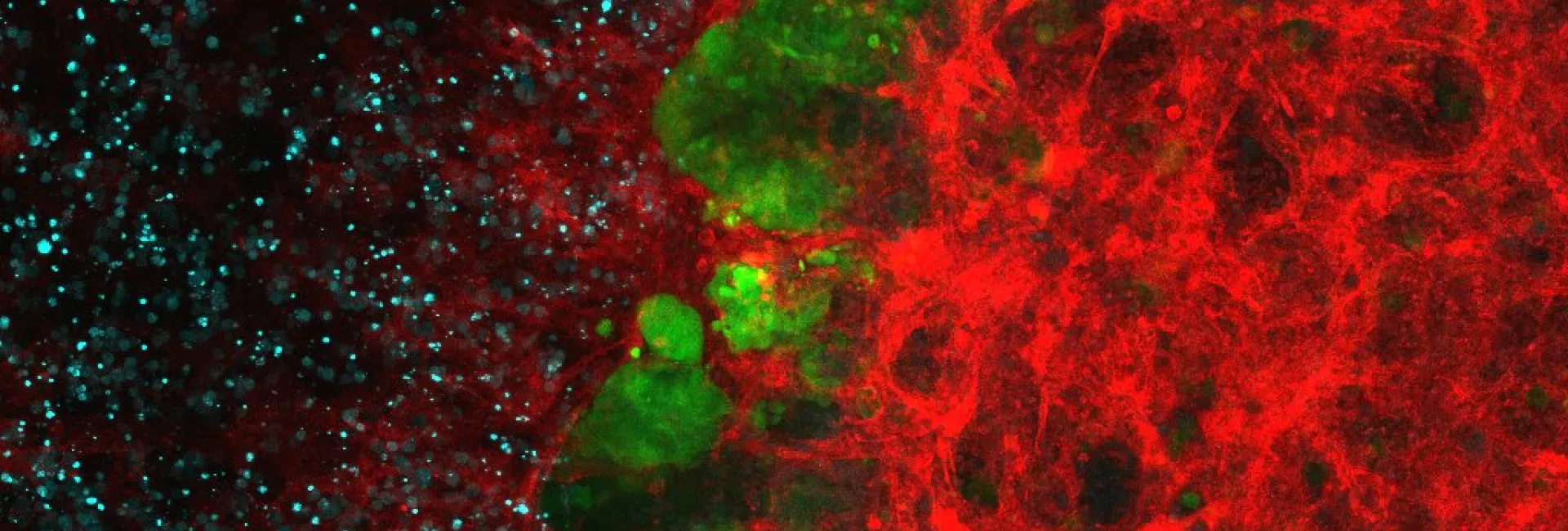
Tumour genomes contain thousands of mutations, which can be identified by Next- Generation Sequencing technologies. By studying the observed pattern of these somatic mutations across genomic regions, we are able to explore the basic cell mechanisms that produce them. The interplay between these mechanisms, such as internal and external insults that damage DNA, chromosomal replication, transcription, and DNA repair mechanisms, leads to mutational processes that give rise to heterogeneous patterns of somatic mutations across the genome.
We have detected, for instance interactions between the machineries of transcription regulation and nucleotide excision repair (NER). Specifically, we recently demonstrated that the binding of transcription factors to their binding sites hinder the efficiency of NER, resulting in the generation of a greater number of mutations in the transcription factor binding site than in neighbouring regions (http://www.nature.com/nature/journal/v532/n7598/abs/nature17661.html).
2. Finding the drivers of cancer
Cancer is mainly a genetic disease. It is caused by genomic alterations that confer somatic cells competitive advantages over neighbouring cells in the same tissue. The genes affected by these alterations are commonly referred to as cancer drivers because they drive the abnormal growth of malignant cells. Recently, important international initiatives have sequenced the exomes and genomes of thousands of tumours belonging to different types of cancer. One of the main goals of the colossal effort channelled into this endeavour is the identification of cancer driver genes and, more recently, also potential driver non-coding genomic elements, such as promoters, enhancers, and non-coding RNAs.
Given that genomic alterations in driver genomic elements are positively selected in the course of tumorigenesis, an effective approach to detect these elements is to find signals of positive selection in their mutational patterns. In recent years, as part of the aforementioned initiatives, we have built bioinformatics tools to identify genomic elements that bear signals of positive selection in their mutational patterns across cohorts of tumours. We call this suite of methods Oncodrives. Using a combination of some of these methods, and others on large pan-cancer cohorts of tumours, we produce comprehensive and reliable catalogues of cancer driver genes. This information is available online at intogen.org
3. Contributing to precision medicine
Precision medicine—understood as the ability to prescribe anti-cancer drugs that specifically target driver alterations in a given tumour—as opposed to traditional chemotherapeutic approaches is an emerging paradigm in clinical and translational research in oncology. The possibility to associate the alterations observed in a patient’s tumour with suitable anti-cancer therapies relies heavily on our ability to accurately identify the alterations that drive the malignancy, as well as those that predict its sensitivity or resistance to drugs.
Our lab seeks to contribute to the advancement of precision medicine, in particular the interpretation of the genomic variants of tumours, thus facilitating the identification of therapeutic options for cancer patients. With this aim, we have developed an approach and several bioinformatics resources to accurately interpret alterations in human tumours. This approach is available at CancerGenomeInterpreter.org.
Selected publications
Projects
“Daño y reparación del ADN en el genoma completo para entender los procesos mutacionales en tumores" (DamReMap), cofinanciado por el Ministerio de Ciencia, Innovación y Universidades- Agencia Estatal de Investigacióny por el Fondo Europeo de Desarrollo Regional (FEDER) de la Unión Europea. Referencia: RTI2018-094095-B-I00



“Finding noncoding cancer drivers" (NONCODRIVERS), financiado por el European Research Council (ERC) mediante el Programa de Investigación e Innovación de la Unión Europea Horizonte 2020. Referencia: 682398

"Exploring mechanisms of resistance in adult and pediatric T-Acute lymphoblastic leukemia", financiado por la Fundación Científica AECC. Referencia: GC10173697BIGA

"Computational ONcology TRaining Alliance" (CONTRA), financiado por la Comisión Europea mediante el Programa de Innovación e Investigación de la Unión Europea Horizonte 2020 bajo la acción Maire Sklodowska-Curie (ITN). Referencia: 766030
Grup de Recerca consolidat (SGR 2017-2019) de la Secretaria d'Universitats i Recerca del Departament d'Empresa i Coneixement de la Generalitat de Catalunya. Agencia de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). Referencia: 2017 SGR 1433


Plataforma de apoyo a la Investigación en Ciencias y Tecnología de la Salud de la convocatoria de 2017 de la Acción Estratégica en Salud 2013-2016, del Instituto de Salud Carlos III (ISCIII), pudiendo estar cofinanciada con cargo al Fondo Europeo de Desarrollo Regional (FEDER). Expediente PT17/0009/0013 (Plataforma de Bioinformática)



Avanzando en la interpretación de genomas de tumores para medicina personalizada del cáncer" (INTERPRETA), cofinanciado por el Ministerio de Economía y Competitividad y por el Fondo Europeo de Desarrollo Regional (FEDER) de la Unión Europea, una manera de hacer Europa. Referencia: SAF2015-66084-R (MINECO/FEDER, UE)


"Creating medically-driven integrative bioinformatics applications focused on oncology, CNS disorders and their comorbidities" (MedBioinformatics), financiado por la Comisión Europea mediante el Programa de Investigación e Innovación de la Unión Europea Horizonte 2020. Referencia: 634143
"Identifying and functionally characterising colorectal cancer driver mutations", financiado por Wellcome Trust (Collaborative Award in Science grant type). Referencia: 214388/Z/18/Z

“Preclinical development of innovative mRNA/MVA vaccines against SARS-CoV2 – COVARNA” con financiación del Departament de Salut – Direcció General de Recerca i Innovació en Salut (DGRIS), de la Generalitat de Catalunya.

"Valorización de EGA para la Industria y la Sociedad”. El proyecto VEIS, con número de expediente 001-P-001647, está cofinanciado en un 50% con 1.951.429,38€ por el Fondo Europeo de Desarrollo Regional de la Unión Europea en el marco del Programa Operativo FEDER de Catalunya 2014-2020, con el soporte del Departamento de Investigación y Universidades. Objetivo: promover el desarrollo tecnológico, la innovación y una Investigación de calidad. El IRB Barcelona recibirá la financiación de 336.551,23€.
“Valorization of EGA for the Industry and Society”. The VEIS project with file number 001-P-001647 has been co-financed by the European Union Regional Development Fund within the framework of the ERDF Operational Program of Catalonia 2014-2020 with a grant of 1,951,429.38€ (50% of total cost eligible) with the support of the Department of Research and Universities. Objective: to promote the technological development, innovation and a high-quality research. IRB Barcelona will receive 336,551.23€ of funding.
“Valorització de EGA per a la Indústria i la Societat". El projecte VEIS amb número d'expedient 001-P-001.647 ha estat cofinançat en un 50%, amb 1.951.429,38 €, pel Fons Europeu de Desenvolupament Regional de la Unió Europea en el marc del Programa Operatiu FEDER de Catalunya 2014-2020, amb el suport del Departament de Recerca i Universitats. Objectiu: promoure el desenvolupament tecnològic, la innovació i una investigació de qualitat. El IRB Barcelona rebrà el finançament de 336.551,23€.


Suport obtingut de l’FSE a través de les Ajuts per a la contractació de personal investigador novell (FI). El Fons Social Europeu dona suport a la creació d’ocupació, ajuda a las persones a aconseguir millors llocs de treball i garanteix oportunitats laborals més justes per a la ciutadania de la Unió Europea.

“iDATA-MP”, cofinanciado por el Instituto de Salud Carlos III y el Fondo Europeo de Desarrollo Regional (FEDER), una manera de hacer Europa, en la convocatoria de infraestructura de Medicina de precisión asociada a la ciencia y tecnología IMPaCT. Referencia: IMP/00019


“Mutaciones somáticas y hematopoyesis clonal como factores de riesgo en aterosclerosis: del laboratorio a la clínica (AtheroClonal)”, proyecto PLEC2021-008194, financiado por MCIN/AEI/10.13039/501100011033 y por la Unión Europea “NextGenerationEU”/PRTR”.
Ayudas Juan de la Cierva-formación 2020, financiado por el Ministerio de Ciencia e Innovación-Agencia Estatal de Investigación y por la Unión Europea“NextGenerationEU”/PRTR. Referencia: IJC2020-044728-I/ MCIN/AEI / 10.13039/501100011033 y por la Unión Europea “NextGenerationEU”/PRTR
“Data-driven cancer genome interpretation for personalised cancer treatment" (CGI-Clinics) financiado por la Comisión Europea mediante el Programa de Innovación e Investigación de la Unión Europea Horizonte Europa. Referencia: 101057509

“Discovering the molecular signatures of cancer PROMotion to INform prevENTion" (PROMINENT) financiado por Cancer Research UK (Referencia: CGCATF-2021/100008), National Cancer Institute (Referencia: 1OT2CA278668-01) y la Asociación Española Contra el Cáncer.
Funded by:


In partnership with:

Grup de Recerca consolidat (SGR-Cat 2021) del Departament de Recerca i Universitats. Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). Referència: 2021 SGR 01480


“Efecto de las quimioterapias en tejidos sanos" CHEMOHEALTH), financiado por MCIN/ AEI /10.13039/501100011033/ y por FEDER Una manera de hacer Europa

"Somatic mutations and clonal hematopoiesis as predictors and drivers of heart failure progression" (MyoClonal), financiado por Fundación "la Caixa" mediante la convocatoria CaixaResearch Health 2022. Referencia: LCF/PR/HR22/52420011
"Discovering the causes of three poorly understood cancers in Europe" (DISCERN), financiado por la Comisión Europea mediante el Programa de Innovación e Investigación de la Unión Europea Horizonte Europa. Referencia: 101096888

Efecto de las quimioterapias en tejidos sanos" CHEMOHEALTH), financiado por MCIN/ AEI /10.13039/501100011033/ y por FEDER Una manera de hacer Europa. Referencia: PID2021-126568OB-I00
“Joint Action Personalised Cancer Medicine (JA PCM)” European Health and Digital Executive Agency. Reference: 101233450

"A European-wide foundation to accelerate Data-driven Cancer Research (EOSC4Cancer)" European Research Executive Agency. Reference: 101058427.

"IOPRED – A disruptive test to predict the response rate of a cancer patient to immunotherapy" Agència de Gestió d’Ajuts Universitaris i de Recerca. Referència: 2022 INNOV 00021.