Advanced Genomic Technologies
The Advanced Genomic Technologies Core Facility strives to empower researchers by offering customized solutions for a wide range of genomic applications, from targeted sequencing to whole-genome studies. We offer a collaborative and flexible approach, including end-to-end support, pilot experiment deployment, comprehensive sample and quality control reporting, and prioritized handling of urgent projects. Our mission is to bring genomics capabilities to every IRB Barcelona research group and external institutions, serving as preferential facilitators of discovery. Reach out to us for details about our portfolio, examples, sample requirements or any other question (genomics@irbbarcelona.org).


The facility provides in-house genome-wide expression and mutation analysis services.
If you are interested in a customized solution not listed below, please contact us at genomics@irbbarcelona.org. We look forward to learning about your projects and working side-by-side with you to overcome any experimental challenge.
If you are ready to submit your samples for analysis, please fill out the appropriate SAMPLE SUBMISSION SHEET found under the ‘Workflow & Forms”’ tab. Alternatively, you are welcome to contact us to schedule an appointment HERE.
- Consultancy and Experimental Design: discuss your project with us and receive information about sample requirements and recommendations for your experimental design. This appointment is highly recommended.
- Quality Control of Extracted Nucleic Acids, Libraries, and Cells: all your samples will be subjected to quality control before processing to inform the best procedures to apply and thereby ensure the highest quality results.
- Nucleic Acid Extraction and Purification: having supported thousands of different projects, we are aware of the many challenges regarding the collection of high-quality DNA and RNA samples. We offer a cost-efficient effective method for RNA extraction from 10-90,000 sorted cells, Drosophila imaginal discs and small soft tissues with less than 100,000 cells. We have obtained high integrity RNA from a single oocyte. We do not provide extraction services for blood, human-derived samples, hard tissues, or samples with more than 100,000 cells, but we will support you with consultancy services.
- Single-Cell Transcriptomics and Multiomics: our single-cell portfolio is diversified and our team has skilled and dedicated staff ready to assist you with:
- Sample Preparation Consultancy: tailored advice and availability for pilots and preliminary QCs to optimize your sample's potential.
- Automated Tissue Dissociation for Single Cell Suspension Preparation: Automated processing of up to 8 tissue samples in the Singleron PythoN for improved consistency and reproducibility.
- Cell Suspension Qualification: quality control of cell suspensions with a LUNA-FL instrument to inform feasibility and assay loading parameters.
- Cell Partitioning and Library Preparation: we employ a variety of advanced technologies to suit any project needs:
- 10x Genomics: versatile solutions for transcriptomic profiling, including GEM-X 3', 5', ATAC, Multiome, and Immune Profiling, and GEM-X Flex for fixed samples.
- Singleron: customizable solutions and automated partitioning using Singleron the Matrix instrument for GEXSCOPE, FOCUSCOPE, and SCIRCLE applications. Ideal for projects with high cell or specific capture requirements.
- Split-seq (e.g. Parse Biosciences): A cost-effective method for high-throughput single-cell RNA sequencing.
- Fast Turnaround Sequencing: opt-in for in-house sequencing and obtain swift and reliable results to feed your draft reviewers. To discuss the viability of your project or to obtain a quote, please email us at genomics@irbbarcelona.org, or schedule a consultation. We ask for a 15-day notice for regular requests, but we accommodate next-day urgent requests for patient-derived samples, subject to personnel and reagent availability.
- Spatial Transcriptomics: as of June 2025 we are deploying in-house spatial transcriptomics capabilities with the 10x genomics Xenium Analyzer, which allows in situ sequencing through cyclic and combinatorial barcoded FISH to quantify gene expression at subcellular resolution in OCT and FFPE tissues.
- Library Preparation for Illumina Next-Generation Sequencing (NGS): we employ fully validated and community-wide accepted procedures for library preparation, thereby ensuring consistent and reliable results. Nonetheless, whenever required, we will align our operating procedures with the specific needs of your project.
- mRNA-, lncRNA-, miRNA-, and sRNA-sequencing (RNA-seq): bulk whole-transcriptome profiling using NGS technologies provides a comprehensive readout of transcript abundance, isoform diversity, as well as the discovery of novel transcripts and fusion genes. Our experienced staff will assist you with sample requisites, workflows, and sequencing strategies to meet the objectives of your project.
- mRNA-seq for drug discovery screens: as of May 2025 we are implementing a cost-efficient and automated procedure to complement drug-screens with a transcriptomics layer of information.
- Whole-Genome Sequencing (WGS): sequencing of the full genome of an individual allows for the identification of somatic or germline DNA variants.
- ChIP-seq, CUT&RUN, and RIP-seq: these approaches combine immunoprecipitation of chromatin (ChIP), or RNA (RIP), with NGS to identify protein-DNA, or protein-RNA, binding sites in a genome-wide manner. In recent years, the facility has supported dozens of ChIP-seq and RIP-seq projects. We will assist you through crosslinking, extraction, and fragmentation procedures to ensure that top quality libraries are obtained.
- Hi-C: the facility has implemented a workflow for chromosome conformation capture and sequencing that allows researchers to map the spatial organisation of genomic DNA within the cell nuclei and identify conformational interactions between non-adjacent regions.
- ATAC-seq: in this case, genome-wide chromatin accessibility is interrogated by transposase accessibility to uncover the extent and identity of regions prone to interact with transcription factors in cancer and other pathologies.
- Targeted Sequencing: while WGS experiments might be your go-to option, their costs can still be prohibitive. Targeted approaches focus throughput, and therefore cost, on defined subsets of genomic regions, thus allowing greater sequencing depths for confident variant detection at lower prices
- Amplicon Sequencing: PCR amplification of specific regions of interest reduces the target genomic space. Several methods are available for a comprehensive and careful panel design, thereby allowing you to multiplex hundreds or thousands of amplicons in a single reaction. Do not hesitate to reach out to obtain a custom design and quote.
- Target Capture, or Sequence Capture and Enrichment: this alternative strategy reduces the sequenced genomic space by “capturing” regions of interest with custom biotinylated probes. Whole-Exome Sequencing is the most widespread Target Capture application. Our staff has extensive experience designing probes for target capture and would be pleased to assist you with your project.
- Automated library preparation with Hamilton Microlab Star: currently available for low input total RNA extractions, mRNA-seq and totalRNAseq procedures.
- Illumina Massively Parallel Sequencing: for outputs of 100, 400, 1.200 and 1.800 Million reads using 50 – 600 effective sequencing cycles in either single-end or paired-end strategies.
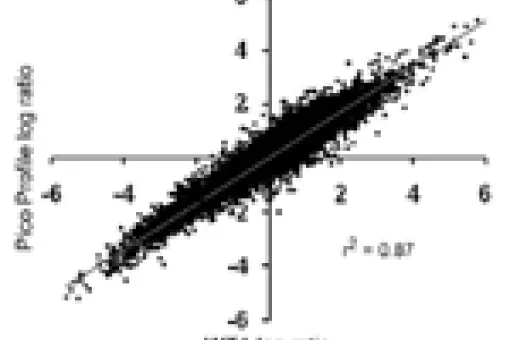
- Pico Profiling: this technique, pioneered by the facility, facilitates the generation of large amounts of saturated cDNA from very small cell populations. Our team has experience isolating RNA and amplifying cDNA from as few as 40 mouse tumour organoid (MTO)-sorted cells or from only one oocyte.
- qPCR: a QuantStudio 6 Pro is available for booking. Runs must start within the working hours of the facility.
- Clone collections: the facility has close to 200,000 clones for your reverse genetics needs, including:
- Human & mouse shRNAs: the complete list of shRNA clones is available in this zip file. Detailed information about the TRC1 clones is available from the Sigma MISSION shRNA Library website and The RNAi Consortium (TRC) based at the Broad Institute.
Human ORF clones: the list of ORF clones is available in this zip file. The sequences of most clones in this collection (release v1.1) can be retrieved from the Harvard Center for Cancer Systems Biology hORFeome v7.1 database.








- Initial Inquiry (service/project qualification): users can schedule an introductory meeting HERE to discuss details of the project, sample characteristics, objectives, etc.
- Quotation sent to the user and PI along with the sample requirements. External users must accept it through signature and PO number.
- Service Scheduling for single cell, spatial transcriptomics, processing of fresh cells, or other services requiring timely coordination between teams.
- Submission of samples and the mandatory service request form with sample details. CF staff reviews the request and, if needed, contacts the user for clarification
- Service processing: samples enter the processing queue and in due time are analysed by facility staff. Waiting times are variable depending on workload and availability, an estimate can be provided at the time of submission.
- Sample quality control: an email with the results of the sample QC will be sent to the user. Qualified samples will continue to downstream processing, non-qualified samples or replicate sets can be replaced by the user.
- Downstream processing and results delivery.
- Follow-up: optional meeting to explain results or assist with interpretation.
SCENTINEL is a collaborative research initiative funded under the European Union's Horizon Europe program, aimed at advancing the capabilities of the Foundation of Research and Technology-Hellas (FORTH) in Greece through the development of genomics and bioinformatics infrastructure using invertebrate tumour models relevant to human diseases. Its goals include enhancing single-cell technology expertise, fostering EU-wide scientific collaborations, and elevating FORTH's global research stature. The project also involves the IRB Barcelona Advanced Genomic Technologies Core Facility, which supports SCENTINEL by facilitating training in single-cell sequencing to ensure the sustainability and advancement of FORTH's research capabilities