Images
The international team achieved it by discovering the rules behind a type of protein structures that are essential for the interaction between proteins and small molecules
IRB Barcelona researcher, Enrique Marcos, is the first author of the study published today in Science
An international team of scientists led by the University of Washington (UW) in Seattle has deciphered key rules that govern how proteins form pocket-like structures that are essential for many key protein functions.
The discovery makes it possible for researchers to custom-design proteins that can mimic the actions of naturally occurring proteins as well as to design new proteins, unlike any found in nature, capable of performing entirely new functions. The results are published today in the January 13 issue of the journal Science.
Currently, scientists hoping to design a new protein to act on particular molecule typically try to repurpose natural proteins, but this strategy is known to have several limitations.
The first author of the study, Enrique Marcos, currently working at Modesto Orozco’s laboratory at the Institute for Research in Biomedicine (IRB Barcelona), explains that “this new strategy will allow us to depend less on natural proteins and to custom-design proteins for medical applications, including the development of new diagnostic tests and treatments, or for biotechnological purposes, such as catalyzing reactions to make industrial processes more efficient”.
David Baker, director of the UW Institute for Protein Design, who led the research, said “This approach will allow scientists to fine tune the size and shape of these pockets or cavities so the custom-designed proteins can bind to and act on specific molecular targets, a process called ligand-binding,” said.
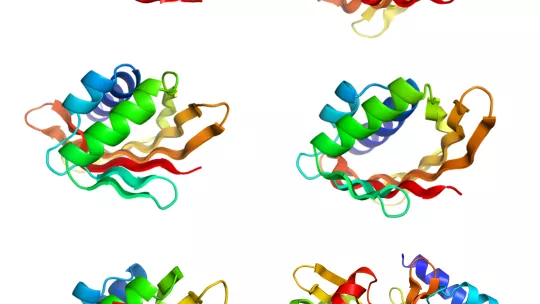
Proteins are made of chains of amino acids that fold into a compact shape that determines their function. In their study, the team studied structures within proteins that are formed when several strands of the protein chain line up next to each other to create sheet-like structures, called β-sheets.
In many natural proteins these sheets bend to form pockets or cavities that bind to target molecules involved in many cellular processes.
To develop the new strategy, the scientists identified the key features in the sequence and orientation of the amino acids that govern how β-sheets flex and curve.
Having identified these features, the researchers show that it is possible to computationally design and experimentally produce with high accuracy protein structures supporting cavities with a variety of shapes and sizes. In addition, they demonstrate that these proteins are highly stable and tolerate high temperatures, which is essential for engineering novel functions.
Research article:
Enrique Marcos, Benjamin Basanta, Tamuka M. Chidyausiku, Yuefeng Tang,Gustav Oberdorfer, Gaohua Liu, G.V.T. Swapna, Rongjin Guan, Daniel-Adriano Silva, Jiayi Dou, Jose Henrique Pereira, Rong Xiao, Banumathi Sankaran, Peter H. Zwart, Gaetano T. Montelione, David Baker
Principles for designing proteins with cavities formed by curved β-sheets
Science (2017). Doi: 10.1126/science.aah7389
About IRB Barcelona
The Institute for Research in Biomedicine (IRB Barcelona) pursues a society free of disease. To this end, it conducts multidisciplinary research of excellence to cure cancer and other diseases linked to ageing. It establishes technology transfer agreements with the pharmaceutical industry and major hospitals to bring research results closer to society, and organises a range of science outreach activities to engage the public in an open dialogue. IRB Barcelona is an international centre that hosts 400 researchers and more than 30 nationalities. Recognised as a Severo Ochoa Centre of Excellence since 2011, IRB Barcelona is a CERCA centre and member of the Barcelona Institute of Science and Technology (BIST).