ERC Advanced Grant, Professor (Biochemistry and Molecular Biology Dept. - UB)

Research information
Research interests
Our research focuses on the study of molecular recognition processes of biological significance from methodological and application points of view. The main subjects addressed are the structure of non-standard nucleic acids, the dynamic properties of macromolecules, and protein-ligand and protein-protein recognition.
Research lines
Methodological aspects
- Analysis of the effect of solvent in molecular recognition. Continuum methods are compared to analyse explicit solvent simulations.
- Fine-tuning of the parametrization of molecular dynamics simulations.
Studies on Small Models
- Theoretical studies of small model systems of large biological impact. This includes, among others, tautomeric and isomerism processes, stacking interactions, salt bridges, hydrogen bonding, cation-π interactions, and general mechanisms of molecular recognition.
Studies on Proteins
- We study the basis of protein interactions, including those of pharmacological importance. In addition, we develop methods to examine both protein-protein and protein-ligand recognition.
- We analyse protein flexibility using massive molecular dynamics simulations.
Studies on Nucleic Acids
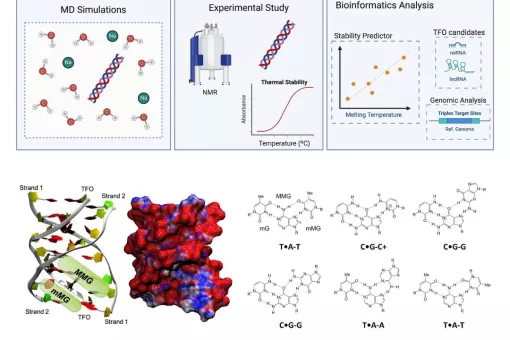
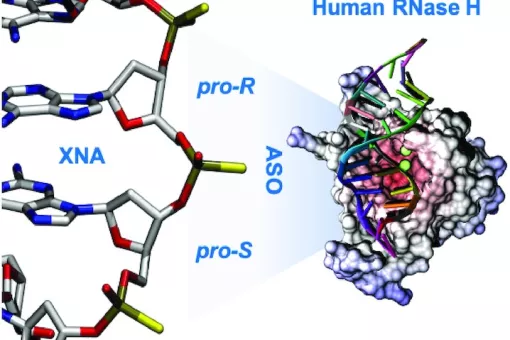
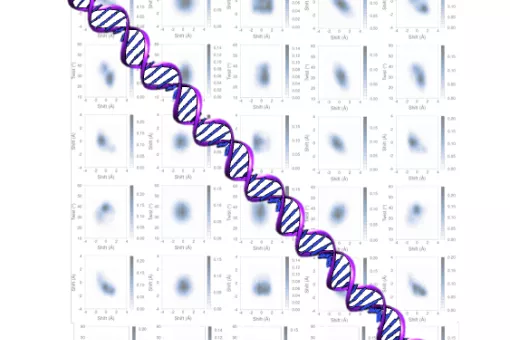
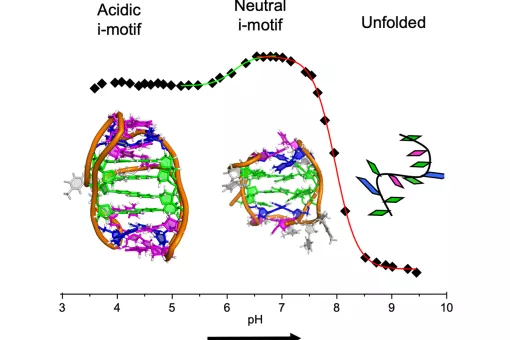
- We use molecular dynamics and statistical mechanics to examine anomalous forms of nucleic acids. Particular attention is paid to the study of conformational transitions in DNA, drug-DNA interactions, and the analysis of triple helices and other structures of potential impact in antigene or antisense therapies.
Bioinformatics
- Data mining of genomic DNA to identify regions with unusual structures or unusual mechanical properties.
- Development of algorithms for the prediction of the pathological character of single nucleotide polymorphisms.
- Structural study of alternative splicing in proteins.
Selected publications
Projects
“Valorización de EGA para la Industria y la Sociedad”. El proyecto VEIS, con número de expediente 001-P-001647, está cofinanciado en un 50% con 1.951.429,38€ por el Fondo Europeo de Desarrollo Regional de la Unión Europea en el marco del Programa Operativo FEDER de Catalunya 2014-2020, con el soporte del Departamento de Investigación y Universidades. Objetivo: promover el desarrollo tecnológico, la innovación y una Investigación de calidad. El IRB Barcelona recibirá la financiación de 336.551,23€.
“Valorization of EGA for the Industry and Society”. The VEIS project with file number 001-P-001647 has been co-financed by the European Union Regional Development Fund within the framework of the ERDF Operational Program of Catalonia 2014-2020 with a grant of 1,951,429.38€ (50% of total cost eligible) with the support of the Department of Research and Universities. Objective: to promote the technological development, innovation and a high-quality research. IRB Barcelona will receive 336,551.23€ of funding.
“Valorització de EGA per a la Indústria i la Societat". El projecte VEIS amb número d'expedient 001-P-001.647 ha estat cofinançat en un 50%, amb 1.951.429,38 €, pel Fons Europeu de Desenvolupament Regional de la Unió Europea en el marc de el Programa Operatiu FEDER de Catalunya 2014-2020, amb el suport del Departament de Recerca i Universitats. Objectiu: promoure el desenvolupament tecnològic, la innovació i una investigació de qualitat. El IRB Barcelona rebrà el finançament de 336.551,23€.
“Cluster Emergente del Cerebro Humano”. El proyecto CECH, con número de referencia 001-P-001682, está cofinanciado en un 50% con 1.527.637,88 € por el Fondo Europeo de Desarrollo Regional de la Unión Europea en el marco del Programa Operativo FEDER Catalunya 2014-2020, con el soporte del Departamento de Investigación y Universidades. Objetivo: promover el desarrollo tecnológico, la innovación y una Investigación de calidad. El IRB Barcelona recibirá la financiación de 219.395,35€
“Emerging cluster of Human Brain”. The CECH project with file number 001-P-001682 has been co-financed by the European Union Regional Development Fund within the framework of the ERDF Operational Program of Catalonia 2014-2020 with a grant of 1,527,637.88 € (50% of total cost eligible) with the support of the Department of Research and Universities. Objective: to promote the technological development, innovation and a high-quality research. IRB Barcelona will receive 219,395.35€ of funding.
"Clúster Emergent del Cervell Humà". El projecte CECH amb número d'expedient 001-P-001682 ha estat cofinançat en un 50%, amb 1.527.637,88 €, pel Fons Europeu de Desenvolupament Regional de la Unió Europea en el marc del Programa Operatiu FEDER de Catalunya 2014-2020, amb el suport del Departament de Recerca i Universitats. Objectiu: promoure el desenvolupament tecnològic, la innovació i una investigació de qualitat. El IRB Barcelona rebrà el finançament de 219.395,35€.
"BioExcel-3 Centre of Excellence for Computational Biomolecular Research". European High-Performance Computing Joint Undertaking (JU). 101093290
“BioExcel-3 Centre of Excellence for Computational Biomolecular Research”, financiado por MCIN/AEI/10.13039/501100011033 y por la Unión Europea “NextGenerationEU”/PRTR”. Referencia PCI2022-135005-2.

Grup de Recerca Consolidat "Modelització Molecular i Bioinformàtica" (SGR-Cat 2021) de l’Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). Referencia 2021 SGR 00863


“Molecular Dynamics Data Bank. The European Repository for Biosimulation Data” financiador por la Comisión Europea mediante el programa Horizon Europe (HORIZON) bajo la acción Developing the European research infrastructures landscape, maintaining global leadership. Referencia 101094651 – MDDB
“Estudio Multiescala del Plegamiento de Ácidos Nucleicos” financiado por el Ministerio de Ciencia e Innovación, perteneciente a la Agencia Estatal de Investigación (AEI). Cofinanciado por el Fondo Europeo de Desarrollo Regional. FEDER, una manera de hacer Europa. Referencia: PID2021-122478NB-I00

“iDATA-MP”, cofinanciado por el Instituto de Salud Carlos III y el Fondo Europeo de Desarrollo Regional (FEDER), una manera de hacer Europa, en la convocatoria de infraestructura de Medicina de precisión asociada a la ciencia y tecnología IMPaCT. Referencia: IMP/00019

“XNA-HUB”, financiado por el Instituto de Salud Carlos III (ISCIII) mediante la convocatoria de Proyectos de I+D+i vinculados a la Medicina Personalizada y Terapias Avanzadas (Iniciativa Transmisiones). Referencia: PMPTA23/00006.