Structural Characterization of Macromolecular Assemblies

ICREA Research Professor
Meet Our Scientists Videos
Research information
Research interests
Our research interests are focused on deciphering the mechanisms that correlate cell signaling with gene expression applying structural biology (NMR, X-ray and SAXS). With our structures, we aim to describe how biological processes occur, how they are regulated and their implications in human diseases.
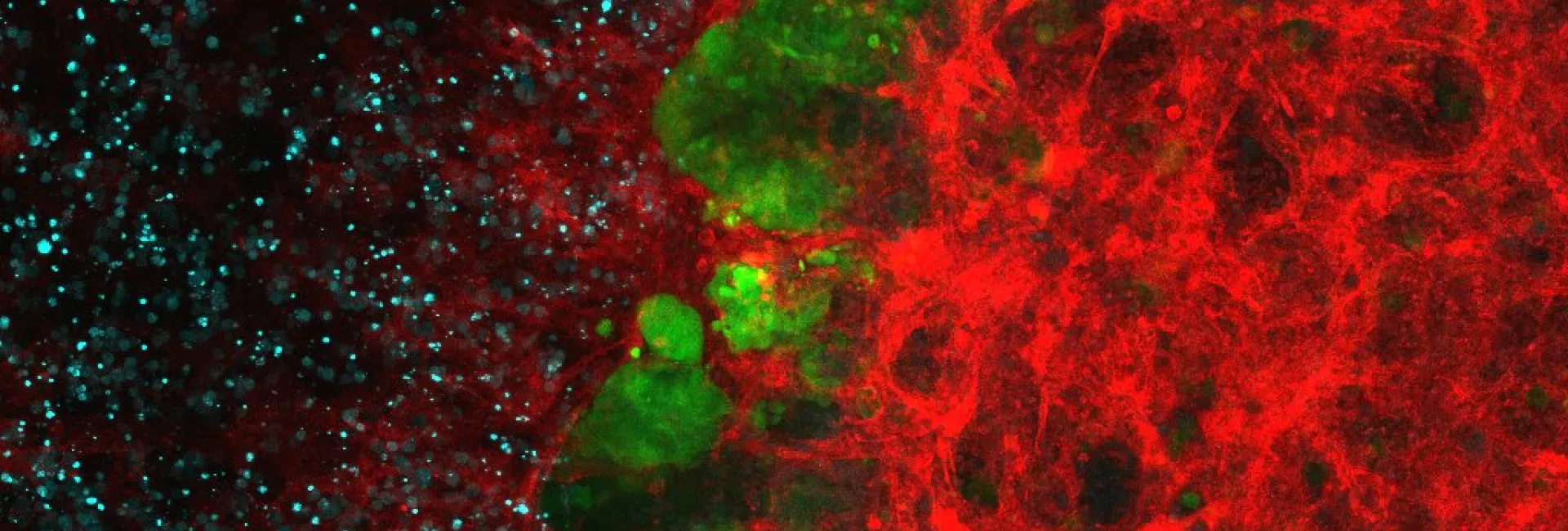
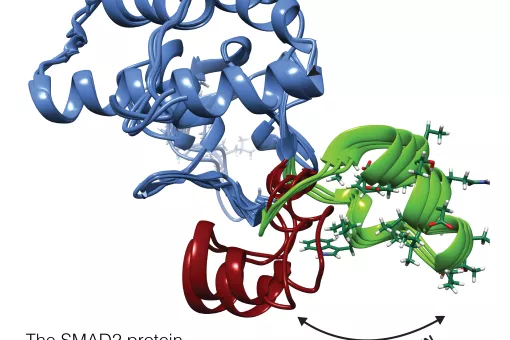
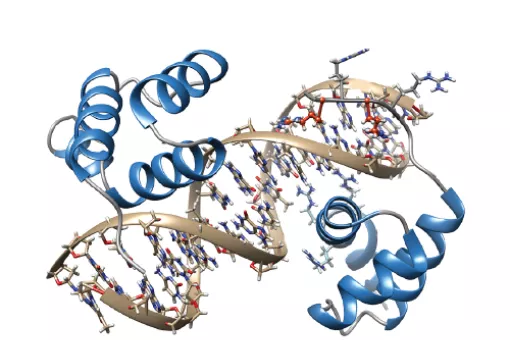
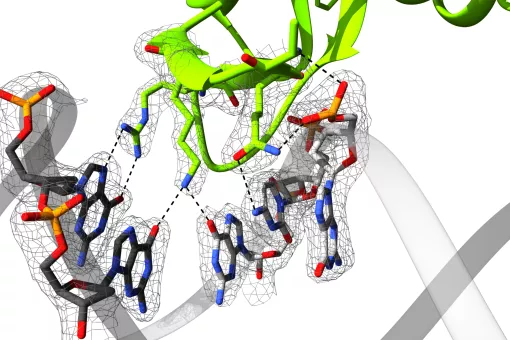
A large part of our work has been devoted to clarifying the role of SMAD transcription factors in TGFβ signaling. SMAD proteins are key components of the TGFβ pathway, one of the four conserved pathways in metazoans. In humans, these proteins are involved in a wide range of cellular responses and are mutated in many oncologic, autoimmune and neurological disorders. Our long-term goal is to explain the complete picture of how full-length SMAD proteins transmit TGFβ signaling and regulate gene expression and cell fate in health and disease.
More recently and in collaboration with medical doctors and researchers of the Sant Pau hospital in Barcelona, we are currently studying the probability of relapse and metastasis of gynecological Cancer patients. We contribute with the analysis of patients’ data from biopsies using artificial intelligence.
Research lines
How Smad proteins and bound cofactors assemble and read DNA
Molecular and structural analysis of ribonucleoprotein complexes
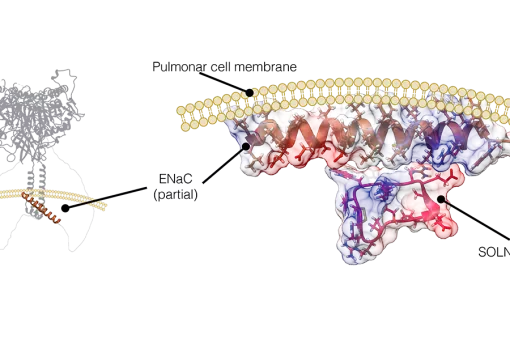
Structural and functional characterization of Somatostatin and Cortistatin analogues
Signatures for the prediction of recurrence and metastasis in Endometrial Cancers.
Computer tools for structural biology
Selected publications
Projects
"Identificación de los sitios de interacción de proteínas Smad con cofactores mediante el uso de biología estructural de alta resolución" (DiSCo-HR), cofinanciado por el Ministerio de Ciencia, Innovación y Universidades- Agencia Estatal de Investigación y por el Fondo Europeo de Desarrollo Regional (FEDER) de la Unión Europea, una manera de hacer Europa. Referencia: BFU2017-82675-P.



"Stem cell differentiation and TGFB-SMAD signaling: Structures of Smad2 and FoxH1 bound to the goosecoid promoter. Analysis of the variants and tumor mutations in these interactions" (MaSSMADs), cofinanciado por el Ministerio de Economía y Competitividad y por el Fondo Europeo de Desarrollo Regional (FEDER) de la Unión Europea. Referencia: BFU2014-53787-P (MINECO/FEDER, UE).


“Indicadors genòmics per a la predicció de la recurrència i la metàstasi en cáncer endometrial”, con el apoyo de la Fundació La Marató de TV3 (201911-30-31).

Suport obtingut de l’FSE a través de les Ajuts per a la contractació de personal investigador novell (FI). El Fons Social Europeu dona suport a la creació d’ocupació, ajuda a las persones a aconseguir millors llocs de treball i garanteix oportunitats laborals més justes per a la ciutadania de la Unió Europea.

“El factor de transcripción Smad4 como diana para el descubrimiento de fármacos: aplicaciones en cancer y enfermedades raras” (Drugs4Smad4), cofinanciado por el Ministerio de Ciencia e Innovación - Agencia Estatal de Investigación (AEI) y por la Unión Europea "NextGenerationEU" mediante el Plan de Recuperación, Transformación y Resiliencia (PRTR). PDC2021-121162-I00/MCIN/AEI/10.13039/501100011033/NextGenerationEU/PRTR
Suport obtingut del SOC pel foment a la contractació en pràctiques de joves en situació d'atur, i finançament al 100% pel Fons Social Europeu com a part de la resposta de la Unió Europea a la pandèmia de COVID-19.

“Complejos SMAD con un papel esencial en el desarrollo y la progresión del cáncer” proyecto PID2021-122909NB-I00 financiado por MCIN/ AEI /10.13039/501100011033/ y por FEDER Una manera de hacer Europa.

"Drugs4RDs -Novel therapeutic strategies against rare diseases", finançat per l’Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) a través de la convocatòria dels ajuts d'Indústria del Coneixement per a l'any 2023 (Llavor i Producte). Referència: 2023 PROD 00095

Síntesi i estructura de biomolècules (SINESBIO)" Grup de Recerca consolidat (SGR-Cat 2021) del Departament de Recerca i Universitats. Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). Referència: 2021 SGR 00866.