Images
- Researchers at IRB Barcelona disclose that the most widely used computational approaches may overlook key information when studying how cells interact within tissues.
- By combining spatial transcriptomics data with data from individual cells in cancer and also in the brain and heart, the study shows that many predicted cell interactions do not correspond with their actual proximity in the tissue.
- The study highlights the need to develop more advanced tools with which to study how cells are coordinated in health and disease.
In a study published in Genome Biology, a team comprising Elena Pareja and Dr. Patrick Aloy at IRB Barcelona has raised questions about the reliability of current methods for predicting intercellular communication. The researchers combined single-cell RNA sequencing data with spatial transcriptomics from cancerous breast tissue, the cerebral cortex, and the heart to systematically analyze how the microenvironment of individual cells influences their genetic activity and interactions with adjacent cells.
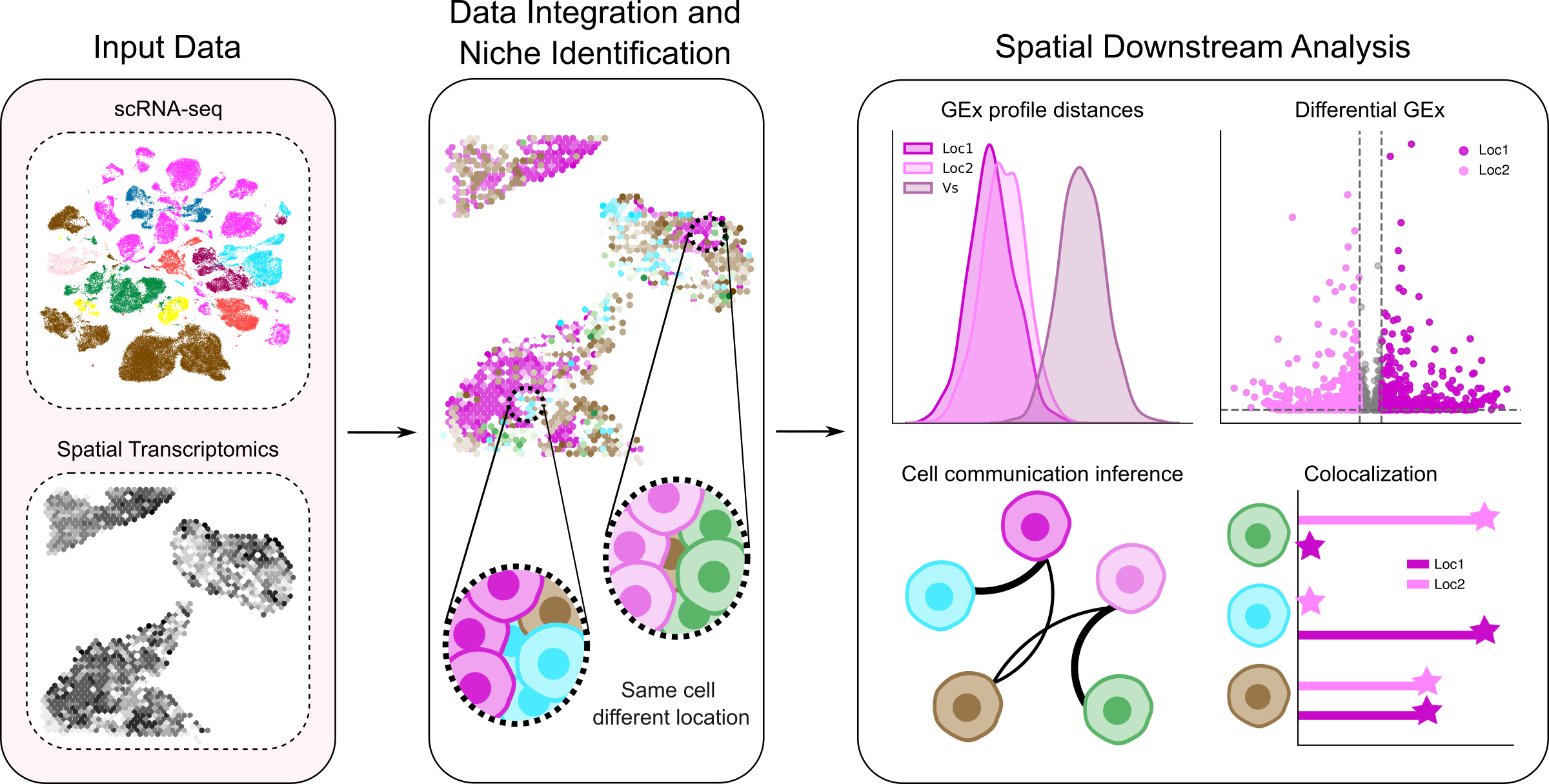
Their analyses revealed that cells of the same type showed no significant differences in their global gene expression, despite being in different tissue contexts. Furthermore, the most popular computational tools used for predicting cell communication—by identifying ligand and receptor genes—frequently indicated interactions between cell types that were not even in close proximity within the tissue.
“These findings are a wake-up call for the field because they show that if we truly want to understand how cells coordinate their functions, we need models that integrate spatial information much more rigorously,” states Dr. Patrick Aloy, ICREA researcher and group leader at IRB Barcelona.
The study suggests that while current methods are useful for proposing potential molecular interactions, they are insufficient to reflect how the local microenvironment dictates actual cell-to-cell dialogue. The authors emphasize the importance of developing new computational approaches that can more accurately identify context-dependent interactions. This is essential for unravelling complex processes such as tissue development, regeneration, and disease progression.
Related article:
Systematic assessment of microenvironment‑dependent transcriptional patterns and intercellular communication
Elena Pareja‑Lorente and Patrick Aloy
Genome Biology (2025) DOI: 10.1186/s13059-025-03677-5
About IRB Barcelona
The Institute for Research in Biomedicine (IRB Barcelona) pursues a society free of disease. To this end, it conducts multidisciplinary research of excellence to cure cancer and other diseases linked to ageing. It establishes technology transfer agreements with the pharmaceutical industry and major hospitals to bring research results closer to society, and organises a range of science outreach activities to engage the public in an open dialogue. IRB Barcelona is an international centre that hosts 400 researchers and more than 30 nationalities. Recognised as a Severo Ochoa Centre of Excellence since 2011, IRB Barcelona is a CERCA centre and member of the Barcelona Institute of Science and Technology (BIST).