Images
- A scientific team at IRB Barcelona has, for the first time, described which hybrid triplexes (DNA-RNA) are most stable within the cell and how they are distributed throughout the human genome.
- The research, published in Nucleic Acids Research, suggests that these triplexes may act as regulatory mechanisms in gene expression and chromatin organization.
- The team has also developed a computational predictor to estimate the stability of various nucleic acid combinations, facilitating future work in molecular biology and epigenetics.
DNA, composed of two complementary strands, contains the genetic information essential for cellular function. For a gene to be activated—or expressed—these strands must separate, allowing part of the sequence to be transcribed into RNA, a process known as transcription.
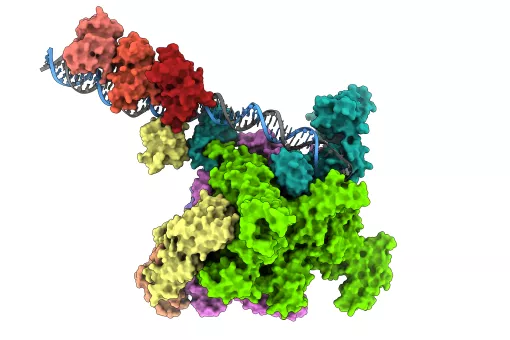
This process is tightly regulated, in part due to special structures that form within DNA or RNA and can modulate gene expression. Among the most notable examples are triplexes, where a third nucleic acid strand binds specifically to the DNA double helix. While DNA-only triplexes have been extensively studied, little has been known until now about those composed of both DNA and RNA—so-called hybrid triplexes—and their biological relevance.
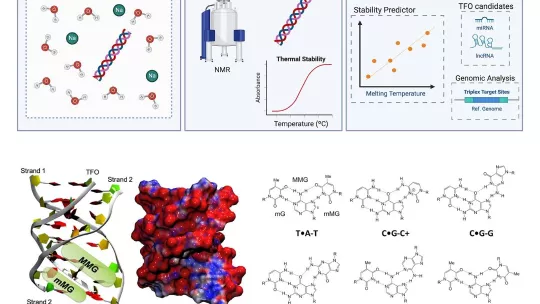
In a recent publication in Nucleic Acids Research, researchers led by Dr. Modesto Orozco at IRB Barcelona, in collaboration with groups from IQAC and IQFBC (CSIC), the University of Barcelona, Nostrum Biodiscovery, and the University of Cambridge, provide a detailed map of the stability and distribution of hybrid triplexes. To carry out the research, the team relied on biophysical experiments, molecular dynamics simulations, and large-scale bioinformatics analyses.
“We wanted to understand which types of hybrid triplexes (DNA-RNA) are more stable, where they tend to form in the genome, and whether they relate to the three-dimensional architecture of DNA. This would help us better understand their potential function as regulatory mechanisms,” explains Dr. Alba Sala, one of the first authors of the study, alongside Dr. Vito Genna, Dr. Guillem Portella, and Dr. Montserrat Terrazas.
After analyzing several DNA-RNA combinations, the researchers discovered that a particular configuration—in which RNA binds to the DNA double helix without fully unwinding it—proved to be the most stable. This finding enabled them to create a computational tool capable of predicting when and where these triplexes might form and whether they could impact gene expression control.
Applying the tool to human genome data revealed that triplexes tend to form in key regulatory regions (such as promoters) and at specific chromatin sites. Thus, the team proposes that these structures may contribute both to the physical organization of DNA and to the activation or repression of various genes.
Implications and Potential Beneficiaries
These findings open up new hypotheses in molecular biology. Researchers in gene regulation and epigenetics may now investigate whether RNA-DNA hybrid triplexes participate in activating or silencing specific genes. Similarly, scientists studying genome architecture could explore how these structures contribute to DNA compaction and stability.
Moreover, biomedical researchers may delve into the presence of abnormal triplexes associated with diseases involving dysregulated gene expression, such as certain cancers.
In the future, identifying these hybrid triplexes could pave the way for new therapeutic strategies, particularly for diseases involving gene regulation malfunctions. Understanding how these triplexes form and stabilize opens exciting possibilities in drug design aimed at correcting defects in the expression of critical genes.
“Our understanding of how RNA regulates chromatin is evolving. These findings provide further evidence that RNA not only serves as a genetic messenger but can also structurally interact with DNA to modulate its function,” comments Dr. Orozco, Head of the Molecular Modelling and Bioinformatics Lab at IRB Barcelona and Professor at the University of Barcelona.
In this study, the team employed thermal stability biophysical experiments, molecular dynamics simulations to visualize strand binding at the atomic level, and exhaustive bioinformatic analyses to locate genomic regions likely to form triplexes.
A Collaborative and Multidisciplinary Effort
This work was made possible through a multidisciplinary collaboration combining experimental and computational techniques, and the contributions of researchers from various specialized centers. It received funding from the European Molecular Biology Organization (EMBO), the Spanish Ministry of Science, the European Research Council (ERC SimDNA), the Platform for Biomolecular and Bioinformatics Resources (ISCIII) co-financed with FEDER funds, and the BioExcel Center of Excellence (H2020) from the European Union.
Artículo de referencia:
Systematic study of hybrid triplex topology and stability suggests a general triplex-mediated regulatory mechanism.
Vito Genna, Guillem Portella, Alba Sala, Montserrat Terrazas, Israel Serrano-Chacón, Javier González, Núria Villegas, Lidia Mateo, Chiara Castellazzi, Mireia Labrador, Anna Aviño, Adam Hospital, Albert Gandioso, Patrick Aloy, Isabelle Brun-Heath, Carlos Gonzalez, Ramon Eritja & Modesto Orozco.
Nucleic Acids Research (2025). DOI: 10.1093/nar/gkaf170
About IRB Barcelona
The Institute for Research in Biomedicine (IRB Barcelona) pursues a society free of disease. To this end, it conducts multidisciplinary research of excellence to cure cancer and other diseases linked to ageing. It establishes technology transfer agreements with the pharmaceutical industry and major hospitals to bring research results closer to society, and organises a range of science outreach activities to engage the public in an open dialogue. IRB Barcelona is an international centre that hosts 400 researchers and more than 30 nationalities. Recognised as a Severo Ochoa Centre of Excellence since 2011, IRB Barcelona is a CERCA centre and member of the Barcelona Institute of Science and Technology (BIST).