Research information
Background
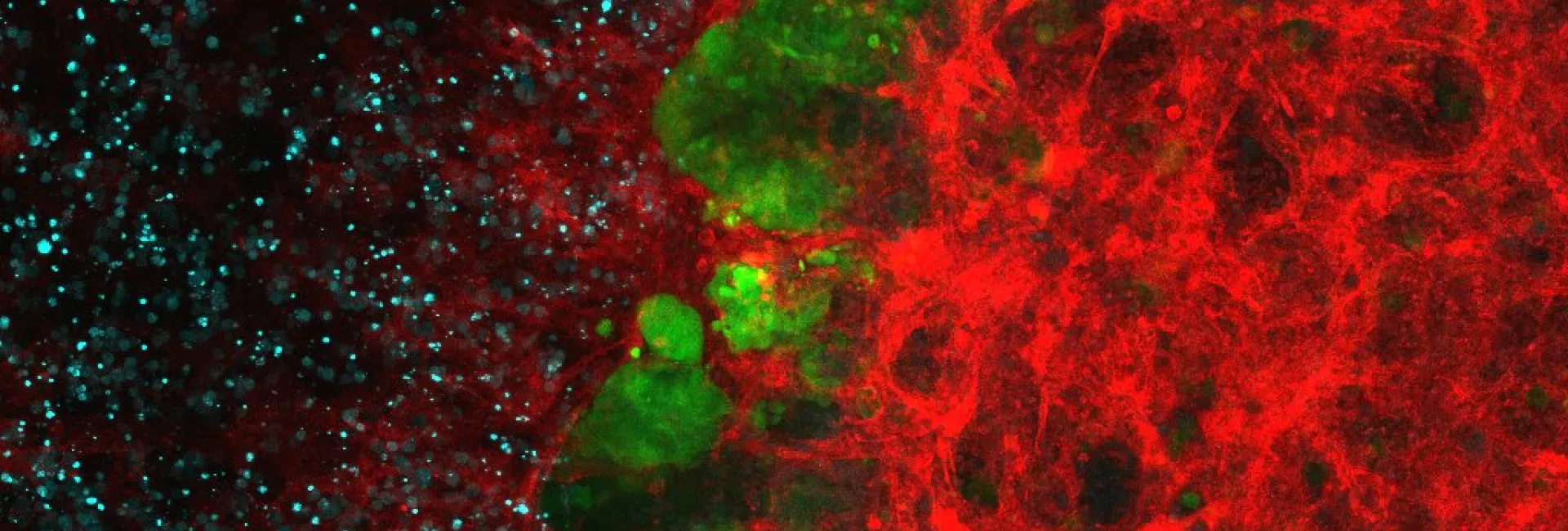
While most cells in our body share almost the same exact genetic information, the way this information is read (the epigenome) is essentially unique in every single cell. If we imagine DNA as a book, then each cell has the same book, but with certain chapters highlighted, some chapters underlined, and others crossed out. This secondary layer of instructions, the epigenome, is what defines whether a cell becomes a brain cell, a skin cell or a blood cell. Importantly, when cells divide, these epigenetic instructions do not get copied with the same fidelity. Thus, as cells proliferate and diversify (during development, or in cancer), their epigenomes drift and vary. Interestingly, some epigenetic information is highly stable and inherited within cellular families (“clones”) defining long-term cellular memory. Other epigenetic information, however, is highly variable, and can be lost or gained within a few cellular divisions. This epigenetic variation underlies the cellular heterogeneity observed in essentially all biological processes, including the topics that we study in our lab: regeneration, immunity, cell plasticity, and tumor therapy resistance.
Research interests
In the lab, we study the topics of stem cell memory and epigenetics in hematopoiesis, immunity and leukemia, with the goal of finding therapies to ultimately have an impact on global public health. Our main experimental approach is to use clonal tracking methods, which allow us to distinguish different cell families within the same sample (tracking in parallel the progeny of thousands of individual cells) and single-cell sequencing, which records tens of thousands of parameters individually in tens of thousands of single cells. Combined, the two technologies are extremely powerful to describe the parameters of regenerative systems, allowing the generation of mathematical models with robust predictive capacity.
First and foremost, we are a technology driven lab, constantly focused on developing the next generation of genetic tools that allow us to answer more complex questions.
We have a basic research goal to understand how cellular diversity emerges, how stem cell memory becomes imprinted, and how these processes (plasticity and memory) impact blood regeneration and cancer.
We also have a translational goal, to use our tools to find markers that predict how cancer cells will respond to a particular treatment, with a focus on understanding the causes of therapy-resistance.
Research lines
Our research lines are centered around these overarching questions:
- How can we record as much biological information as possible in a complex multicellular system without perturbing the organism?
- How are the cellular dynamics of blood regeneration controlled? How does individual epigenetic variation influence these dynamics? How does it impact aging, chronic inflammation and cancer?
- How is stem cell memory formed during development and in response to injury? How does it self-perpetuate?
- How do leukemic cells become therapy-resistant? How can we make leukemic cells more sensitive to immunotherapy?
Selected publications
Projects
"Mecanismos moleculares de la resistencia a terapia y plasticidad celular de la leucemia mieloide aguda" (AMLTRAP), financiado por MCIN/ AEI/10.13039/501100011033. Referencia: PID2020-114638RA-I00

"Origins and Consequences of Hematopoietic Stem Cell Memories" (MemOriStem), financiado por el European Research Council (ERC) mediante el Programa de Innovación e Investigación de la Unión Europea Horizonte Europa. Referencia: 101042992

“Systems analysis of therapy-resistance in acute myeloid leukemia” (SYSTIC), financiado por Fundación CRIS de Investigación para Vencer el Cáncer (CRIS contra el Cáncer). Referencia: PR_EX_2020-24

Grup de Recerca emergent (SGR-Cat 2021) del Departament de Recerca i Universitats. Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). Referència: 2021 SGR 01586


"Funding of £2000 approved for Scientific Meeting and funding of £500 approved for Sustainable Conferencing," entitled, European Developmental Biology Congress: Barcelona 2023 to take place on 27/09/2023. Funded by THE COMPANY OF BIOLOGISTS LIMITED

"Una plataforma de ensayos de perturbación paralelizados con secuenciación de células individuales para identificar susceptibilidades epigenéticas en células de leucemia (MUSICAL)". Ministerio de Ciencia, Innovación y Universidades. Reference: PDC2025-166663-I00