Research information
Research interests
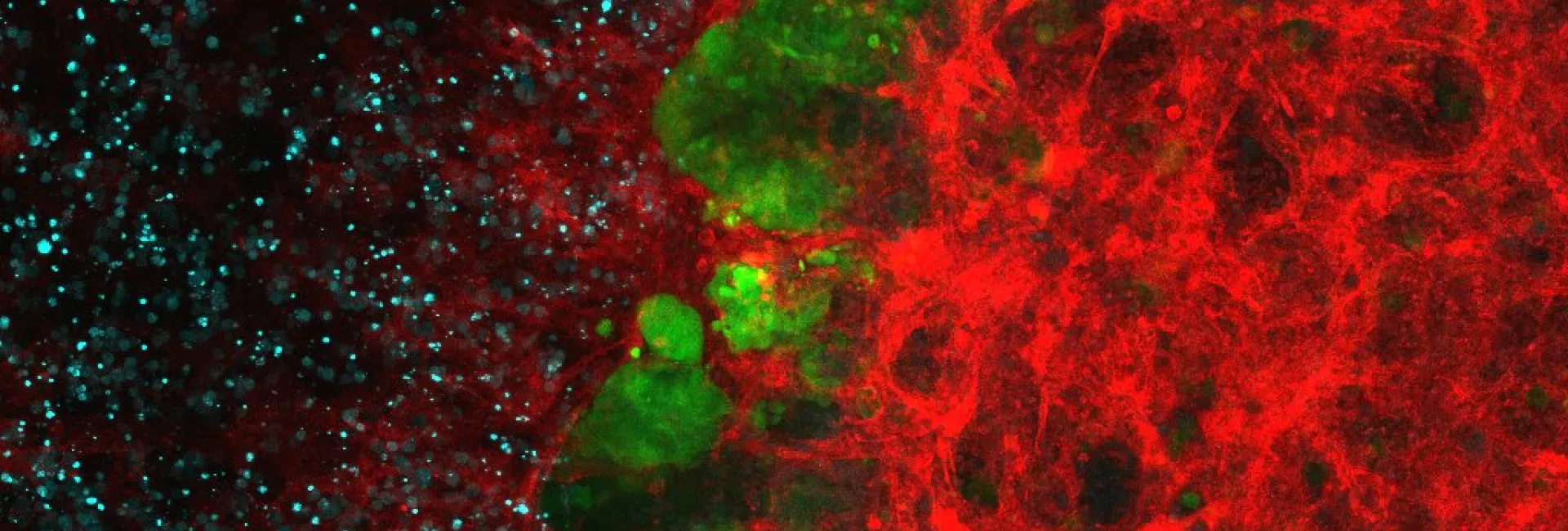
We aim to unravel how cells detect and respond to environmental changes. We focus our studies on the characterisation of stress signal transduction pathways, especially those regulated by MAP kinases of the Hog1/p38 family, also known as the stress-activated MAP kinases (SAPKs). Proper adaptation to stress involves the modulation of several basic aspects of cell biology, among them the cell cycle and gene expression. Using S. cerevisiae budding yeast as a model organism, as well as higher eukaryotic cells, we are dissecting the molecular mechanisms underlying cell response to changes in the extracellular environment and characterising the adaptive responses required for cell survival. Based on our knowledge of signal transduction and using synthetic biology, we also seek to modify cell behaviour to reprogram cell response to specific inputs/stimuli.
Research lines
1. SAPK signalling: Using quantitative data in single cells and mathematical modelling, together with mutational analyses, we study the basic signalling properties of stress-responsive MAP pathways and how to alter them.
2. SAPK targets: Using proteomics, biochemistry and genetics, our main goal is to identify new targets for SAPKs and thus widen our understanding of cellular adaptation to stress. This information is expected to facilitate the characterisation of the bases of adaptation in eukaryotes.
3. Cell cycle control: SAPKs act in several phases of the cell cycle to allow prompt response to extracellular stimuli and the maintenance of cell integrity. We are uncovering the mechanisms by which Hog1 and p38 SAPKs regulate the cell cycle.
4. Regulation of mRNA biogenesis: SAPKs control critical steps of mRNA biogenesis and are thus key regulators of stress-responsive gene expression. Our main aim is to determine the contribution of multiple factors to overall gene expression in response to stress. We are also using genome-wide CRISPR screening to identify essential genes for stress adaptation.
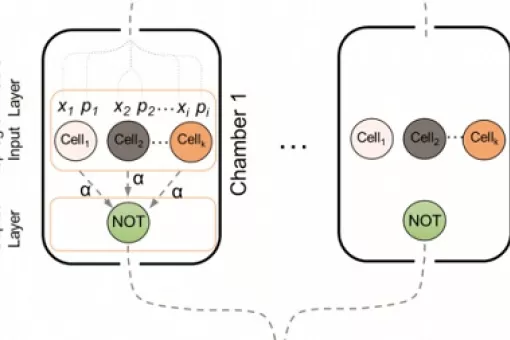
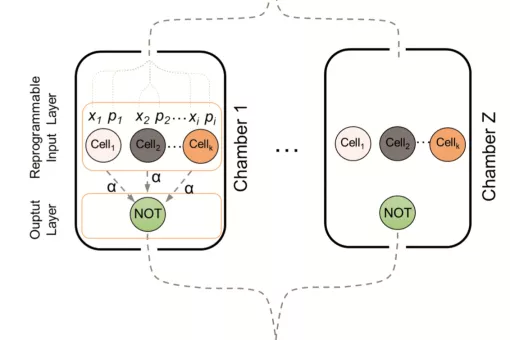
5. Synthetic biology: We are interested in implementing complex engineered networks to perform in vivo cellular computation. In a joint effort between theoretical and experimental groups, we have established biological circuits with distributed computation. We are now addressing the application of cellular computation to relevant health issues, such as diabetes.
Selected publications
Projects
"Regulación transcripcional mediante modificaciones de histonas" Ministerio de Ciencia, Innovación y Universidades and Fondo Europeo de Desarrollo Regional (FEDER-UE). Reference: BFU2017-85152-P.



“Regulation of retinoblastoma (Rb) as a new therapeutic strategy for the treatment of breast cancer” Asociación Española contra el Cancer (AECC). Reference: PROYE18010POSA

"Circuits cel·lulars sintètics encapsulats per restablir control glicèmic en diabetis mellitus tipus 1" Fundació La Marató de TV3. Reference 201610.31

Grup de Recerca consolidat (SGR 2017-2019) Secretaria d'Universitats i Recerca del Departament d'Empresa i Coneixement de la Generalitat de Catalunya. Agencia de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). Reference: 2017 SGR 779


“Disección de la Adaptación al Estrés en Eucariotas” Ministerio de Ciencia, Innovación y Universidades and Fondo Europeo de Desarrollo Regional (FEDER). Reference: PGC2018-094136-B-100



"Characterization of the impact of stress responses in cellular physiology" Ministerio de Ciencia e Innovación/Fondo Europeo de Desarrollo Regional (FEDER). Reference: PID2021-124723NB-C21

"Consorcios biológicos reprogramables como dispositivos de detección múltiple de contaminantes ambientales (BioCont)" Ministerio de Ciencia e Innovación. NextGenerationEU/PRTR. Reference: TED2021-131562B-I00

"Pequeñas moléculas moduladoras de la actividad de RB para el tratamiento del cáncer" Ministerio de Ciencia e Innovación. NextGenerationEU/PRTR. Reference: PDC2022-133807-I00

"Harnessing an alternative RB1 activation mechanism for a potential breast cancer therapy". Asociación Española contra el Cancer (AECC). Reference: PRYGN247170POSA

"Decodificando principios conservados de la adaptación al estrés (DecoAdapt)". Ministerio de Ciencia, Innovación y Universidades/Fondo Europeo de Desarrollo Regional (FEDER). Reference: PID2024-156607NB-C21

"Supresión de la resistencia a los inhibidores de CDK4,6 en el tratemiento del cáncer mediante la reprogramación de la actividad de RB1 (ReBoot)". Ministerio de Ciencia, Innovación y Universidades. Reference: PDC2025-165528-I00

Group members
Group members
What are we doing now?
We’re thrilled to announce that Ivan has successfully defended their PhD thesis. Well done Ivan!!!
We enjoyed a well-deserved lab retreat with great weather, views, and food!! We had some wonderful days filled with great science, fun, and bonding.
Join our research group as a postdoctoral fellow and study the regulation of the Retinoblastoma (RB) protein as a new therapeutic strategy for cancer. You will explore how a specific RB mutant can override CDK activity in normal and tumor cells and mouse models.
Your expertize and leadership will undoubtedly propel the institution to greater heights. We are excited to see the positive impact you'll make.
His work focused on the identification and characterization of LRRC8A chloride channel in cell volume regulation and adaptation to osmotic shock.