Structural Biology of Protein & Nucleic Acid Complexes and Molecular Machines

Professor (IBMB-CSIC)
Research information
Background
Our research focuses on the three-dimensional structure of proteins, nucleic acids and their complexes with the aim to further our understanding of several essential mechanisms in the cell. We use a number of molecular biology and structural biology techniques, with a focus on X-ray diffraction crystallography. Our approach usually starts with the cloning of relevant genes and the expression and purification of the encoded proteins. Proteins, nucleic acids and their complexes are then crystallized and analysed by X-ray diffraction, using synchrotron radiation. The final outcome is a detailed 3D view of the molecular structures at atomic resolution.
Research interests
We examine systems related to horizontal gene transfer which involve the DNA translocation across the cell membranes. In addition, we address the regulatory mechanisms of gene expression and the control mechanisms of DNA replication. We also study unique DNA structures, like DNA junctions, and novel drugs that target DNA.
Research lines
DNA replication control. Plasmids are extra-chromosomal DNA molecules that replicate autonomously. In pathogenic bacteria, plasmids may carry antibiotic resistance genes. Plasmid replication is controlled by gene products encoded in the plasmid itself. In collaboration with Manuel Espinosa and Gloria del Solar (CIB-CSIC), we are currently studying this system in streptococcal plasmids.
Horizontal gene transfer. Conjugation is the main route for horizontal gene transfer in bacteria and is responsible for the spread of antibiotic resistance. During conjugation, plasmid DNA is processed and transported across cell membranes between cells. The relaxosome is a protein-DNA complex formed by several proteins and a DNA segment called the Origin of Transfer. The transport complex is a transmembrane multiprotein assembly. In collaboration with Fernando de la Cruz (U. of Cantabria), we are examining this system in E. coli plasmid R388.
DNA packaging in viruses. Portal proteins or connectors are large multimeric proteins involved in DNA packaging into viral capsids. With the help of other proteins, they translocate the DNA through one vertex of the capsid. We are currently analysing various connectors and other related DNA packaging proteins, in collaboration with José L. Carrsacosa and José María Valpuesta (CNB-CSIC).
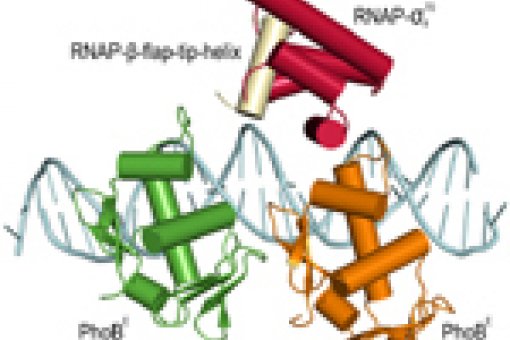
Transcription regulation. We study several transcription factors and their complexes with other proteins and DNA promoter regions. In collaboration with Margarita Salas (CBM-CSIC), in bacteriophage φ29 we examine the transcriptional regulator p4 that functions as a switch between early and late gene expression during the infection cycle. In E.coli, we address the PhoB transcriptional activator, a response regulator of the two-component signal transduction system that controls the expression of more than 40 genes related to phosphate assimilation.
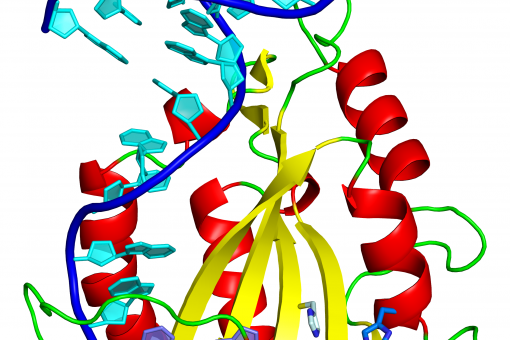
DNA structure and drug-DNA interactions. Unique DNA structures, such as four-way and three-way junctions related to DNA recombination and other processes, are being structurally analysed. Novel DNA-binding drugs that target these peculiar structures and other sequence-specific drugs are also under study.
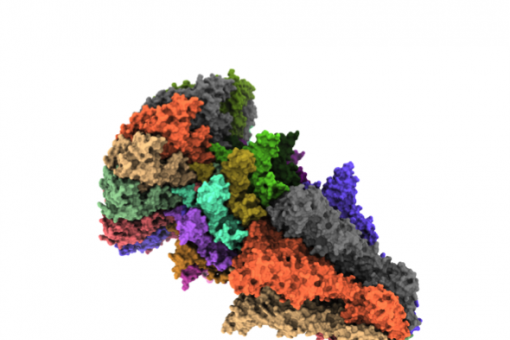
Viral replication machinery. Within the Structural Genomics European Consortium VIZIER, the replication enzymes and ancillary proteins of human pathogenic RNA virus are currently being structurally characterized. The final aim is to provide tools for the discovery of new antiviral agents.
Protein complexes, epigenetic regulation of gene expression and cancer. As a partner of the European Consortium 3D-REPERTOIRE and coordinator of the Spanish Structural Genomics Consortium GENES, our group studies several protein complexes and molecular machines, some of which are related to the epigenetic control of gene expression. Targets for anticancer therapy are among the proteins currently being analysed.